| Contents |
|
| General | Back to top |
WebScipio offers an advanced option to search for exons of tandem gene duplications. It is based on the exon-intron structure determined by Scipio. The algorithm of the search divides into several steps, which are conducted for each known exon. It takes three assumptions into account: Firstly, duplicated exons have a similar length; secondly, their splice sites and reading frames are conserved; thirdly, they are homologues. Several parameters of the search are adjustable.
| Options | Back to top |
Allowed length difference for exons
Determines the maximal difference between the length of an exon and the length of the duplicated exon. The length is defined by the number of amino acids coded by the exon.
Minimal score for exons
The score is defined by the global alignment score of the translated exon sequence to the duplicated exon translation divided by the global alignment score of the exon translation to itself. The minimal score is a threshold above which the duplicated exon is taken into account.
Minimal tandem gene score
The score is defined by number of amino acids of the original gene found in the duplicated gene divided by the number of amino acids of the original gene. This means that mismatches are counted, but gaps are not counted.
Minimal exon length
Just exons, which are longer than the minimal exon length, are taken into account for the search for tandem gene duplications. The length is defined by the number of amino acids coded by the exon.
Search for concatenated exons
If this option is enabled, the algorithm searches for duplicated exons with pairs (2-tuples) of neighbouring exons, triplets (3-tuples), 4-tuples, 5-tuples and so on up to all exons by concatenating the translations of the neighbouring exons. So helps to find duplicated exons in which introns are lost.
Search for splitted exons
If this option is enabled, the algorithm searches for exons splitted in two exons in the duplicated gene. Only exons, which were not found in the first round of the algorithm are used for this search. The missing exons are splitted in all possible two parts, whereby the smaller part must be longer than the minimal exon length.
Search with start codon for first exon
The first exon does not have a conserved splice site pattern at the 5' end but its beginning is determined by the start codon (ATG) coding for a Methionin. So the algorithm tries to find duplicated exons beginning with ATG (and not with a splice site pattern at the 5' end of the exon).
- Auto: This option makes it possible to autodetect, whether the first exon in the Scipio result is the beginning of the protein. It searches for duplicated exons with the ATG start codon (instead of a splice site pattern) if the first exon begins at the start of the protein sequence and starts with ATG.
- Enable: This option forces the algorithm to look for alternative exons with ATG as start codon for the first exon (and does not use the splice site pattern).
- Disable: This option forces the algorithm to use splice site patterns of the first exon to look for duplicated exons as if it is a normal exon in the middle.
Search with stop codons for last exon
The last exon does not have a splice site pattern at the 3' end but its end is determined by a stop codon (TAG, TAA, TGA). So the algorithm tries to find duplicated exons followed by a stop codon (and not with a splice site pattern at the 3' end of the exon).
- Auto: This option makes it possible to autodetect, whether the last exon in the Scipio result is the end of the protein. It searches for duplicated exons with stop codons (instead of a splice site pattern) if the last exon ends at the end of the protein sequence and is followed by TAG, TAA or TGA.
- Enable: This option forces the algorithm to look for duplicated exons followed by stop codons for the last exon (and does not use the splice site pattern).
- Disable: This option forces the algorithm to use splice site patterns of the last exon to look for duplicated exons as if it is a normal exon in the middle.
| Examples | Back to top |
Oreochromis niloticus mysion heavy chain 13 (Mhc13) gene with two tandem gene duplications
| Organism | Oreochromis niloticus |
| Genome file | |
| Query sequence | MNDTDMEVFGVAAPYLRKSERERIAAQNVPFDAKSAVFVPHPKQEYVKGKIRSQDGTKVN VEIEDGKVVTVHVDDIRPMNPPKFDKIEDMALLTHLHEPAVLFNLKERYAAWMIYTYSGL FCVTVNPYKWLPVYNPEVVAGYRGKKRQEAPPHIFSISDNAYQYMLTDRENQSILITGES GAGKTVNTKRVIQYFATITAMGESSKKEQLGSKMQGTLEDQIIQANPLLEAFGNAKTVRN DNSSRFGKFIRIHFGTKGKLASADIETYLLEKSRVTFQLLAERSYHIFYQILSNKKPDLI EMLLITSNPYDYPFISQGEITVLSINDAEELMASDRAIDILGFSTEEKVGIYKLTGAVMH NGNMKFKQKQREEQAEPDGTEVADKVAYLMGLNSADLLKALCCPRVKVGNEYVTKGQTPQ QVNNAVGALSKAVYEKLFLWMVTRINQQLDTKLPRQHFIGVLDIAGFEIFEINSLEQLCI NFTNEKLQQFFNHHMFVLEQEEYKKEGIEWEFIDFGMDLAACIELIEKPMGIFSILEEEC MFPKATDGSFKNKLYDQHLGKNSIFQKPKPSKAKTEAHFSLMHYAGTVDYNISGWLEKNK DPLNDTVVQLYQKASLKLLCQLFATYASADAAADGNKKNYKKKGSSFQTVSALFRENLNK LMANLRSTHPHFVRCIIPNETKIPGIMDHHLVLHQLRCNGVLEGIRICRKGFPSRILYGD FRQRYRILNASVIPEGQFIDSKKASEKLLSSIDVDHTQYRFGYTKVFFKAGLLGLLEEMR DERLAVLMTRIQAVARGYVTRLRLKEMMKKREAVYIIQYNIRSFMNVKNWPWMKLFFKIK PLLRSAEAEKEMQTMKEEFARLREELAKSEARRKELEEKMVMLMQEKNDLYLQIQAEREN LCDAEERCEGLIKSKIHLEAKAKEFSERMEEEEEINAEITAKKRKLEDECSELKRDIDDL ELTIAKVEKEKYATENKVKNLIEELTTLEENLLKSSKEMKALQEVHQQTLDDLQAEEDRV NSLIKTKTKLEQQIDDLEGLVEQEKKLRADLERSRRKLEGDLKLNQETIMDLENERQQAE EQLKKKDFEISSLQSKIDDEQALSTQLQKKIKELQARTEELEEEIEAERAARAKVEKQRS DLSRELEEITERLEEAGGASAAQAELNKKREAEIQRLRHELEESTLQHESIAVALRKKQA DSVAELGDQIENLQRVKQKLEKEKSEMKMEIDDMARSMETVLKSKANVEKQCRSLEDQMN EYKTKADEAQRSLSDYTTLSARLQTENGELTRLLEEKESILSQVNRGKTAAGHKIEEMKR LLDEEIKTKNALAHSLQSSRHDCELLREQYEEEQEAKAELQRCLSKVNSDVAQWRNKYET DTIQRTEELEEAKKKLVQRLQESEEMTEAANVKCASLEKTKQRLQAEVEDLMVELERSNA ANATLDKKQKNFDKVLAEWKQKYEECQSDLEVSQRESRALNTELFKLKNSYEEVLDHLES MKRDNKNLQQEITDINEQVGESTKMLRELEKATKHAEQEKRDTQAALEEAESSLEQEESK ILQLELELNQIKSEVERKVAEKDEEIDQLKRNYQRTVDYLQSTLDAETRSRNDAVRMKKK MEGDLNEMEIQLGHANRQAAEATKQLRNLQTQLKDTQVHLDEALQRQEDLKEELAIIERR NNLMMAENEELRASLEQSDRSRKLAEQELMEVSERVQLLHSQNTSLVNSKKKMEADLTQL QSEMEETMQEARNADEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNLEATVKDLQQRL DEAEQVALKGGKKEIQKLEAKVRELENELEAEQKRSGEAVKGVRKYERKIKELTYQGEEE KKNSARLQDLVNKLQLKVKAYKRQFEEAEEQSSIHLAKFRKVQHELEEAEERADVAESQL NKLRARSRDVAGKGELKS |
| YAML-file for upload | |
| Expert options | defaults, except "Region Size = 50000" |
| Search for tandem gene duplications | defaults, except "Minimal score for exons = 30" and "Region Size = 50000" Enable search! |
| Result |

|
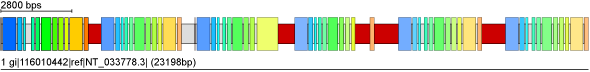
Drosophila melanogaster CG30047 gene with five tandem gene duplications
| Organism | Drosophila melanogaster |
| Genome file | |
| YAML-file for upload | |
| Query sequence | MKSREKNGSAASNSDVALVNVLSQQKLRRHRLPWYYAPSFLLLWVALFYAVVYPLYHRLP DSVLISHESSKPGQFVAERAQRLLYKYDKIGPKVVGSVANEVTTVAFLEEEVENIRAAMR SDLYELQLDVQHPSGAYMHWQMVNMYQGVTNVVVKISSRSSNSSSYLLVNSHFDSKPSSP GSGDDGTMVVVMLEVLRQVAISDTPFEHPIVFLFNGAEENPLEASHGFITQHKWAGNCKA LINLEVAGSGGRDLLFQSGPNNPWLIKYYYQNAKHPFATTMAEEIFQSGILPSDTDFRIF RDYGQLPGLDMAQISNGYVYHTIFDNVQAVPIDSLQSSGDNALSLVRAFADAPEMQNPED HSEGHAVFFDYLGLFFVYYTENTGIVLNCCIAVASLVLVVCSLLRMGRESDVSIGRVSIW FAIILVLHVLGMILSLGLPLLMAVLFDAGDRSMTYFSNNWLVIGLFIVPAIIGQILPLTL YYTLKPNDEISHPNHIHMSLHAHCVLLSLIAIILTAISLRTPYLCMMSLLFYGGALLINL LSTLHDRGYYWVVLVQVLQLVPFLYFCYLFYTFLLVFFPMLGRFGHGTNPDLLIALICAV GTFFALGFVAPLINIFRWPKLALLGLGVVTFIFSMIAVSEVGFPYRAKTSVMRIHFLHVR RIFYEYDGSVSLSDSGYYFDFQDRRLYYPLENTSVNLTGLASTSSECDKYLMCGVPCFNH RWCKTRAKSHWLPREQEVAIPGATSLKLLSKAVLDSGKVARFEFEISGPPHMSLYIQPLD GVEVEDWSFIRNMLDEPDTYSPPYQIFFAYGADNTPLKFHIDFAKSSGDFSTPTFQLGFA ASFVSYDYDRDAAGLKFISDFPDFAHVMEWPTLYERYIF |
| Expert options | defaults, except "BLAT Tilesize = 5" |
| Search for tandem gene duplications | defaults, except "Search for concatenated exons = enabled" and "Allowed length difference for exons = 5" Enable search! |
| Result |
|
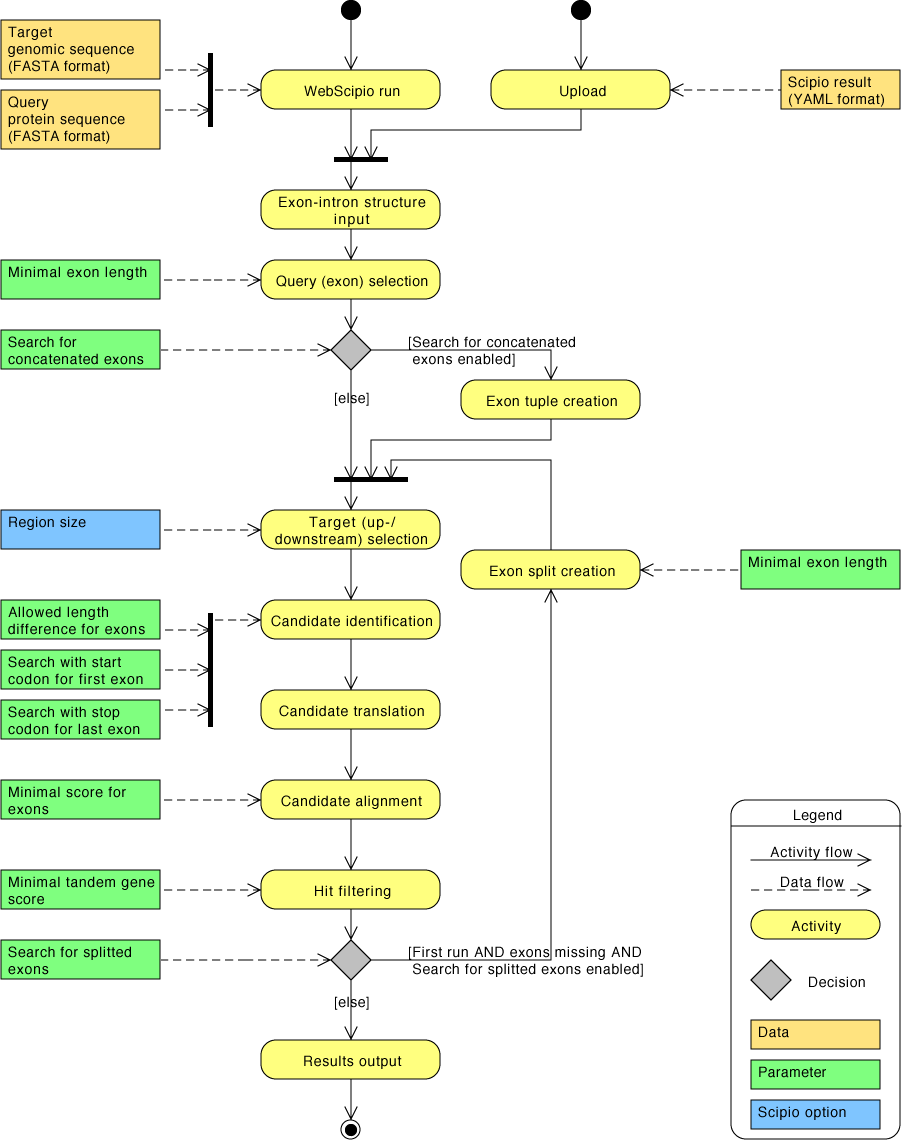
| Overview of search parameters as activity diagram | Back to top |
|